Abstract
Evaluation of a 113-gene predictor of benefit from neoadjuvant T-AC treatment in an I-SPY2 cohort
Background:
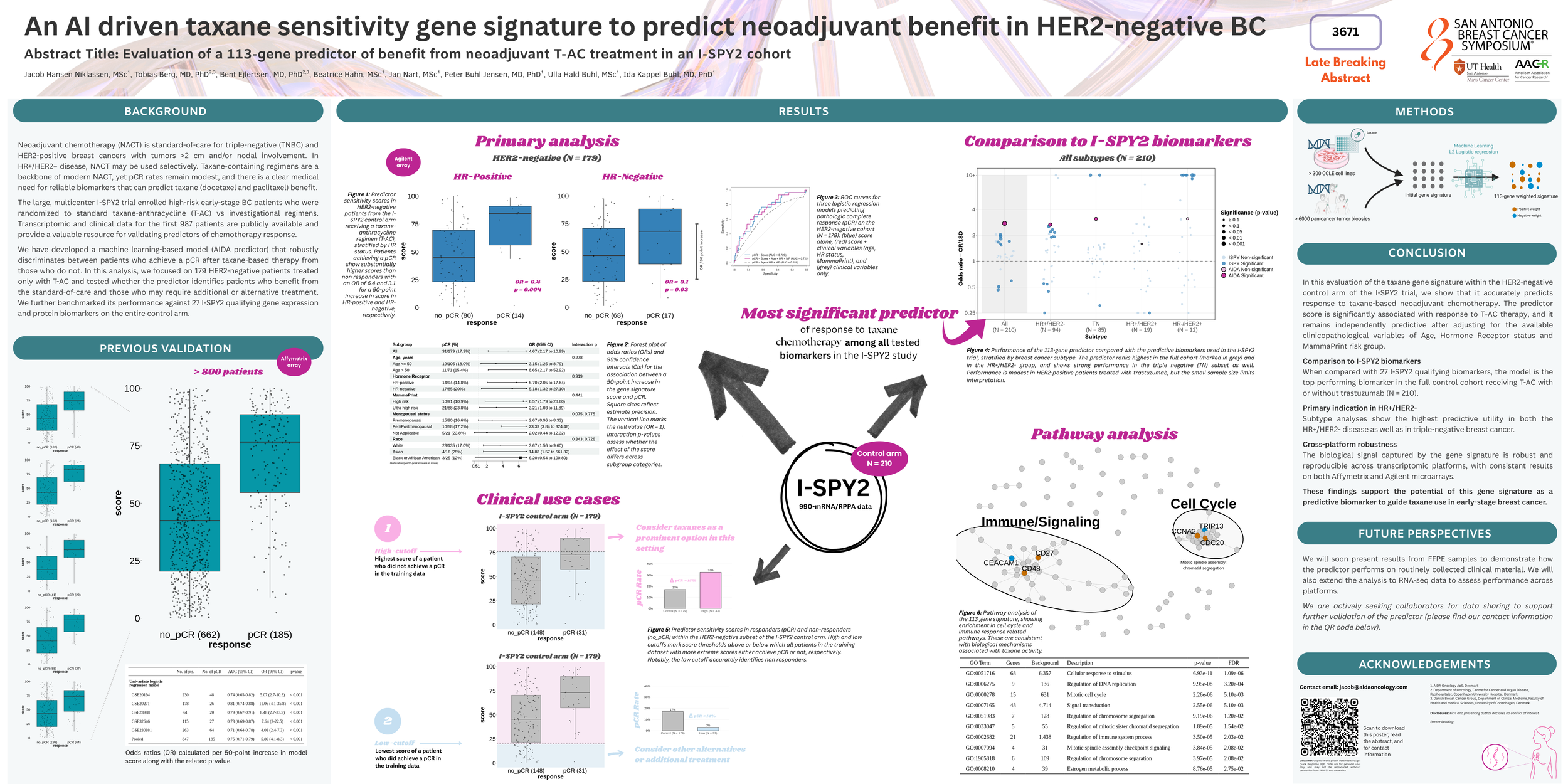
Neoadjuvant chemotherapy is standard in early-stage breast cancer (BC), enabling tumor downstaging and treatment tailoring. Taxane-based regimens are widely used in both hormone receptor (HR)-positive/HER2-negative and triple-negative BC, yet pathological complete response (pCR) rates remain modest. In the I-SPY2 trial, high-risk early-stage BC patients were randomized to standard taxane-anthracycline (T-AC) vs investigational regimens. We demonstrate that within the shared HER2-negative control arm, a 113-gene expression-based machine learning model can stratify patients by likelihood of benefit from T-AC, supporting biomarker-driven strategies to improve pCR.
The 113-gene model, which was developed using data from T-FEC treated patients analyzed on the Affymetrix HG U133A and HG U133 Plus 2.0 arrays, has previously been tested in five independent cohorts (GEO IDs: GSE20194, GSE20271, GSE23988, GSE32646, GSE230881) and shown to discriminate patients achieving pCR to taxane-based therapy from those who do not (published at ESMO 2025). Here, we present further validation of this 113-gene signature in data from the I-SPY2-990 mRNA Data Resource and demonstrate its applicability to Agilent microarrays in addition to Affymetrix.
Methods:
The previously developed model was adapted to the Agilent platform, where 108 of the 113 genes were available, and then applied to gene expression data from 179 high-risk, early-stage BC pre-treatment tumor samples, from patients who were treated with T-AC and evaluated for pCR at time of surgery. Patients were stratified based on HR status and scored on a scale from 0-100. The association of patient score (per 50-point increase) with pCR was assessed using logistic regression models.
Results:
In this analysis of 179 high-risk, early-stage, HER2-negative patients from a phase II prospective study, with gene expression data from the Agilent 4x44k microarray, we evaluated a 113-gene machine learning-based predictor of response to taxane-based regimens. The model, originally built on data from Affymetrix microarrays and previously evaluated on five external cohorts, was able to successfully distinguish patients with pCR from those without pCR after neoadjuvant T-AC treatment. The association between model score and treatment outcome was significant in both univariate and multivariable settings. These findings provide further validation of the predictor and support its robustness across different microarray platforms.
Conclusions:
In this analysis of 179 high-risk, early-stage, HER2-negative patients from a phase II prospective study, with gene expression data from the Agilent 4x44k microarray, we evaluated a 113-gene machine learning-based predictor of response to taxane-based regimens. The model, originally built on data from Affymetrix microarrays and previously evaluated on five external cohorts, was able to successfully distinguish patients with pCR from those without pCR after neoadjuvant T-AC treatment. The association between model score and treatment outcome was significant in both univariate and multivariable settings. These findings provide further validation of the predictor and support its robustness across different microarray platforms.
| Dataset / subtype | N | Obtained pCR | Response Rate | OR | P value |
|---|---|---|---|---|---|
| GSE20194 | 230 | 48 | 20.9% | 5.07 | <0.001 |
| GSE20271 | 178 | 26 | 14.6% | 11.06 | <0.001 |
| GSE23988 | 61 | 20 | 32.8% | 8.48 | <0.001 |
| GSE32646 | 115 | 27 | 23.5% | 7.64 | <0.001 |
| GSE230881 | 263 | 64 | 24.3% | 4.08 | <0.001 |
| I-SPY2 (GSE194040) | 179 | 31 | 17.3% | 4.50 | <0.001 |
| HR-negative | 85 | 17 | 20% | 3.37 | 0.02 |
| HR-positive | 94 | 14 | 14.9% | 5.70 | 0.006 |
Authors and affiliations:
Jacob Hansen Niklassen: Niklassen, J.H. (1), Tobias Berg: Berg (2, 3), Bent Ejlertsen: Ejlertsen, B. (2, 3), Beatrice Hahn: Hahn, B. (1), Jan Nart: Nart, J. (1), Peter Buhl Jensen: Jensen, P.B. (1), Ulla Hald Buhl: Buhl, U.H. (1), Ida Kappel Buhl: Buhl, I.K. (1)
1: Aida Oncology ApS, Denmark
2: Department of Oncology, Centrefor Cancer and Organ Disease, Rigshospitalet, Copenhagen University Hospital, Denmark
3: Danish Breast Cancer Group, Department of Clinical Medicine, Faculty of Health and medical Sciences, University of Copenhagen, Denmark
Contact
Jacob Hansen Niklassen: jacob@aidaoncology.com